Python NumPy to Elixir-Nx
Learn how to leverage existing codebases from the Python ML ecosystem
For my ElixirConfUS talk, I wanted to demonstrate training a spam detection model with in Elixir. A pre-processing step I needed to perform was TF-IDF vectorization, but there were no TF-IDF libraries already written which were built for Elixir-Nx. Seeing as TF-IDF is an extremely common pre-processing step with Decision Trees, since they ingest tabular data, I decided to go ahead and write a full-fledged implementation rather than just writing a minimal implementation that I needed for my example.
I decided to model my implementation after the TF-IDF Vectorizer implementation in the Python scikit-learn, and in writing it I learned many lessons about translating Python NumPy code to Elixir-Nx. Since Nx is to Elixir as NumPy is to Python, I thought others might find it useful to see how we can leverage existing code from the Python ecosystem to bring it Elixir.
The sklearn source code can be found here, while the Elixir source code can be found here.
API Overview
A primary goal I had while writing my Elixir library was to make the API as similar to the Python API as possible so that it would be easy for people coming from Python, since sklearn is the Machine Learning library that most people likely have experience with. The Elixir version supports most of the same arguments as the sklearn version. Here are some examples of basic usage of the API
# Elixir
TfidfVectorizer.new(
ngram_range: {1, 3},
sublinear_tf: true,
stop_words: english_stop_words,
max_features: 5000
)
|> TfidfVectorizer.fit_transform(X_train)# Python
TfidfVectorizer(
sublinear_tf=True,
ngram_range=(1, 3),
max_features=5000
).fit_transform(X_train)Design Overview
We will start by looking at the Sklearn CountVectorizer class since TfidfFVectorizer is actually implemented as a subclass of CountVectorizer and then using the fit method of the TfidfTransformer class to transform the output of the CountVectorizer to its TF-IDF representation. As a result, the bulk of the code is implemented in the CountVectorizer. In Elixir, I accomplished this design by having a CountVectorizer module and a TfidfVectorizer module which has a CountVectorizer as a struct member.
The vectorizer works by building a vocabulary from the given corpus (or using a vocabulary you pass it), counting the number of times each word in the vocabulary is taken, and filtering according to
The vectorizers work roughly according to these steps:
- Either builds a vocabulary from the given corpus or uses a vocabulary supplied to it.
a. Performs preprocessing according to a given function
b. Performs tokenization according to a tokenization function
c. Generates requested ngrams
d. Filters stop words - Iterates through each document in the corpus, counting each term in the vocabulary
- Limit output features according to parameters
a. max_features - Only consider the topmax_featuresordered by term frequency across the corpus.
b. min_df - Ignore terms that have a document frequency strictly lower than the given threshold.
c. Ignore terms that have a document frequency strictly higher than the given threshold. - Output
CountVectorizer TfidfVectorizeruses theCountVectorizerto transform the term-frequency matrix into it TFIDF Matrix.
As you can see, the bulk of the work is done in the CountVectorizer module, so that is where we will spend the most of our time going forward. Now that you have a general understanding of how the vectorizers work, we will look at a brief survey of different functions to compare Python and Elixir implementations.
Implementation Details
Here is how the vectorizer is initialized in Python. It uses keyword args with default parameters and initializes its class attributes accordingly.
class CountVectorizer(_VectorizerMixin, BaseEstimator):
def __init__(
self,
*,
input="content",
encoding="utf-8",
decode_error="strict",
strip_accents=None,
lowercase=True,
preprocessor=None,
tokenizer=None,
stop_words=None,
token_pattern=r"(?u)\b\w\w+\b",
ngram_range=(1, 1),
analyzer="word",
max_df=1.0,
min_df=1,
max_features=None,
vocabulary=None,
binary=False,
dtype=np.int64,
)
self.input = input
self.encoding = encoding
self.decode_error = decode_error
self.strip_accents = strip_accents
self.preprocessor = preprocessor
self.tokenizer = tokenizer
self.analyzer = analyzer
self.lowercase = lowercase
self.token_pattern = token_pattern
self.stop_words = stop_words
self.max_df = max_df
self.min_df = min_df
self.max_features = max_features
self.ngram_range = ngram_range
self.vocabulary = vocabulary
self.binary = binary
self.dtype = dtypeAnd here is a snippet of the CountVectorizer struct in Elixir as well as the new/1 function that is used to initialize the vectorizer. new/1 gives a similar behavior to Python's class init method. I used NimbleOptions for parameter validation (it's a great library and you can read more about it here), and you can refer to the parameters source here. validate_shared! validates the parameters and assigns default value when none were provided.
defmodule Mighty.Preprocessing.CountVectorizer do
defstruct vocabulary: nil,
ngram_range: {1, 1},
max_features: nil,
min_df: 1,
max_df: 1.0,
stop_words: [],
binary: false,
preprocessor: nil,
tokenizer: nil,
pruned: nil
def new(opts \\ []) do
opts = Mighty.Preprocessing.Shared.validate_shared!(opts)
struct(__MODULE__, opts)
end
endAll of the operations in our CountVectorizer module will take a CountVectorizer as its first argument, which allows us to pipe our operations nicely. The Python implementation separates the TfidfTransformer from the TfidfVectorizer, where the TFIDFVectorizer inherits from the CountVectorizer. To achieve similar behavior, our TfidfVectorizer is its own struct that contains a CountVectorizer as one of its member. Creating a new TfidfVectorizer starts with creating a new CountVectorizer:
defmodule Mighty.Preprocessing.TfidfVectorizer do
alias Mighty.Preprocessing.CountVectorizer
alias Mighty.Preprocessing.Shared
defstruct [
:count_vectorizer,
:norm,
:idf,
use_idf: true,
smooth_idf: true,
sublinear_tf: false
]
@doc """
Creates a new `TfidfVectorizer` struct with the given options.
Returns the new vectorizer.
"""
def new(opts \\ []) do
{general_opts, tfidf_opts} =
Keyword.split(opts, Shared.get_vectorizer_schema() |> Keyword.keys())
count_vectorizer = CountVectorizer.new(general_opts)
tfidf_opts = Shared.validate_tfidf!(tfidf_opts)
%__MODULE__{count_vectorizer: count_vectorizer}
|> struct(tfidf_opts)
end
endNow, let's compare three of the main pieces of functionality between the two implementations: building the term-frequency matrix (count matrix), limiting / pruning features from the resulting matrix, and performing the TFIDF transformation on that resulting matrix.
Building Term-Frequency Matrix
defp _transform(vectorizer = %__MODULE__{}, corpus, n_doc) do
if is_nil(vectorizer.vocabulary) do
raise "CountVectorizer must be fit to a corpus before transforming the corpus. Use CountVectorizer.fit/2 or CountVectorizer.fit_transform/2 to fit the CountVectorizer to a corpus."
end
tf = Nx.broadcast(0, {n_doc, Enum.count(vectorizer.vocabulary)})
corpus
|> Enum.with_index()
|> Enum.chunk_every(2000)
|> Enum.reduce(tf, fn chunk, acc ->
Task.async_stream(
chunk,
fn {doc, doc_idx} ->
doc
|> then(&do_process(vectorizer, &1))
|> Enum.reduce(
Map.new(vectorizer.vocabulary, fn {k, _} -> {k, 0} end),
fn token, acc ->
Map.update(acc, token, 1, &(&1 + 1))
end
)
|> Enum.map(fn {k, v} ->
case Map.get(vectorizer.vocabulary, k) do
nil -> nil
_ when v == 0 -> nil
idx -> [doc_idx, idx, v]
end
end)
end,
timeout: :infinity
)
|> Enum.reduce({[], []}, fn
{:ok, iter_result}, acc ->
Enum.reduce(iter_result, acc, fn
nil, acc -> acc
[x, y, z], {idx, upd} -> {[[x, y] | idx], [z | upd]}
end)
end)
|> then(fn {idx, upd} ->
Nx.indexed_put(acc, Nx.tensor(idx), Nx.tensor(upd))
end)
end)
end
def _count_vocab(self, raw_documents, fixed_vocab):
"""Create sparse feature matrix, and vocabulary where fixed_vocab=False"""
if fixed_vocab:
vocabulary = self.vocabulary_
else:
# Add a new value when a new vocabulary item is seen
vocabulary = defaultdict()
vocabulary.default_factory = vocabulary.__len__
analyze = self.build_analyzer()
j_indices = []
indptr = []
values = _make_int_array()
indptr.append(0)
for doc in raw_documents:
feature_counter = {}
for feature in analyze(doc):
try:
feature_idx = vocabulary[feature]
if feature_idx not in feature_counter:
feature_counter[feature_idx] = 1
else:
feature_counter[feature_idx] += 1
except KeyError:
# Ignore out-of-vocabulary items for fixed_vocab=True
continue
j_indices.extend(feature_counter.keys())
values.extend(feature_counter.values())
indptr.append(len(j_indices))
if not fixed_vocab:
# disable defaultdict behaviour
vocabulary = dict(vocabulary)
if not vocabulary:
raise ValueError(
"empty vocabulary; perhaps the documents only contain stop words"
)
if indptr[-1] > np.iinfo(np.int32).max: # = 2**31 - 1
if _IS_32BIT:
raise ValueError(
(
"sparse CSR array has {} non-zero "
"elements and requires 64 bit indexing, "
"which is unsupported with 32 bit Python."
).format(indptr[-1])
)
indices_dtype = np.int64
else:
indices_dtype = np.int32
j_indices = np.asarray(j_indices, dtype=indices_dtype)
indptr = np.asarray(indptr, dtype=indices_dtype)
values = np.frombuffer(values, dtype=np.intc)
X = sp.csr_matrix(
(values, j_indices, indptr),
shape=(len(indptr) - 1, len(vocabulary)),
dtype=self.dtype,
)
X.sort_indices()
return vocabulary, X
The most evident differences here are that in the Elixir code we are building a dense tensor, while the Python code is building a sparse tensor. A sparse tensor certainly makes much more sense in the context of these vectorizers, but as of now Nx currently does not support sparse tensors. This is also why we are using Task.async_stream along with Enum.chunk_every, as to reduce the memory consumption since it is dense.
The way we are constructing the tensor, however, is almost identical. We start by creating a zero-tensor the size of the final tensor. Then we are creating mappings of indices and their updates during our iteration within the reduce. After we collect these updates, we update the initial zero-tensor using Nx.indexed_put, which requires a list of indices you are updating along with the new values you are putting into those indices.
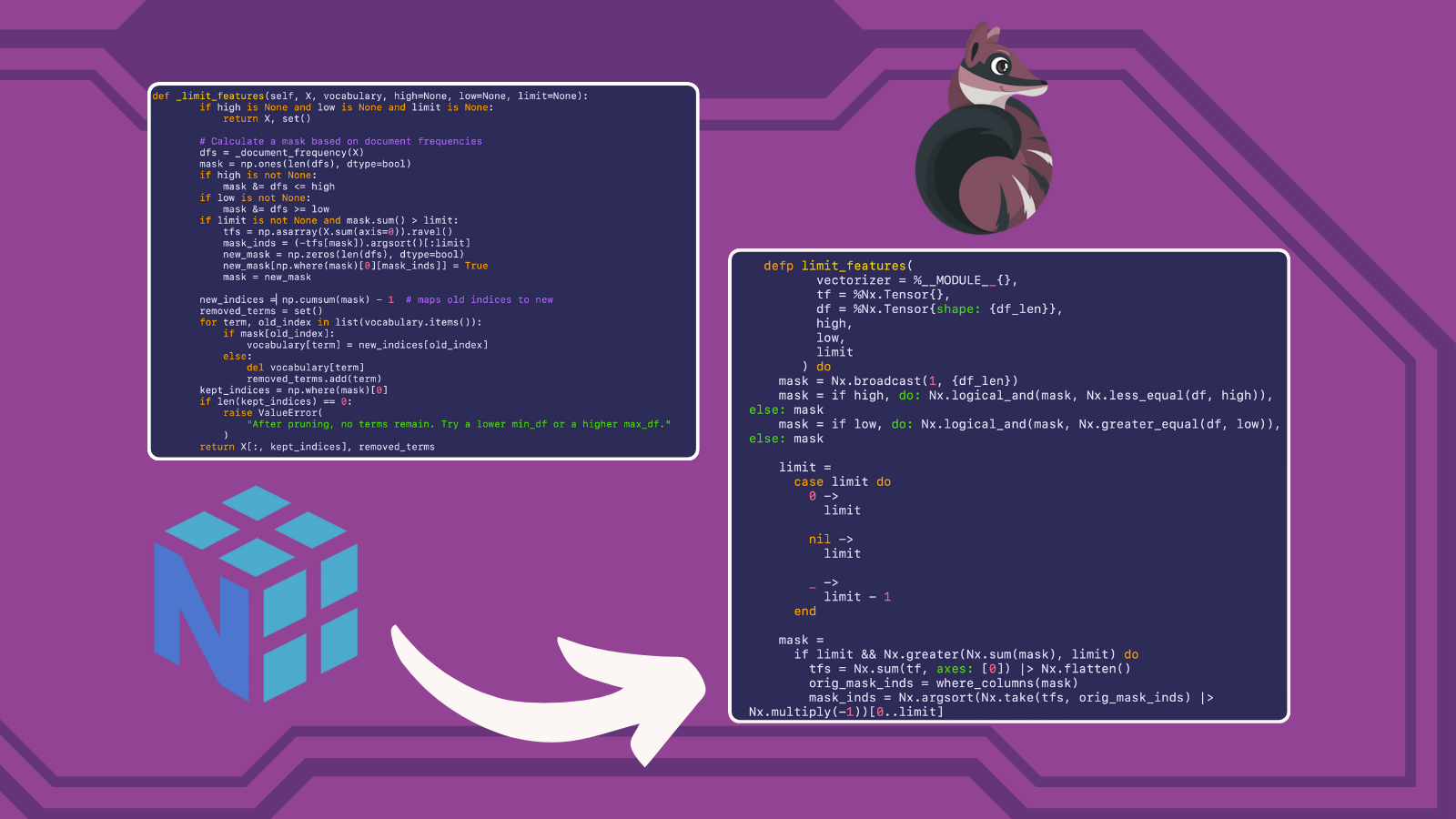
Feature Pruning
defp where_columns(condition = %Nx.Tensor{shape: {_cond_len}}) do
count = Nx.sum(condition) |> Nx.to_number()
Nx.argsort(condition, direction: :desc) |> Nx.slice_along_axis(0, count, axis: 0)
end
defp limit_features(
vectorizer = %__MODULE__{},
tf = %Nx.Tensor{},
df = %Nx.Tensor{shape: {df_len}},
high,
low,
limit
) do
mask = Nx.broadcast(1, {df_len})
mask = if high, do: Nx.logical_and(mask, Nx.less_equal(df, high)), else: mask
mask = if low, do: Nx.logical_and(mask, Nx.greater_equal(df, low)), else: mask
limit =
case limit do
0 ->
limit
nil ->
limit
_ ->
limit - 1
end
mask =
if limit && Nx.greater(Nx.sum(mask), limit) do
tfs = Nx.sum(tf, axes: [0]) |> Nx.flatten()
orig_mask_inds = where_columns(mask)
mask_inds = Nx.argsort(Nx.take(tfs, orig_mask_inds) |> Nx.multiply(-1))[0..limit]
new_mask = Nx.broadcast(0, {df_len})
new_indices = Nx.take(orig_mask_inds, mask_inds) |> Nx.new_axis(1)
new_updates = Nx.broadcast(1, {Nx.flat_size(new_indices)})
new_mask = Nx.indexed_put(new_mask, new_indices, new_updates)
new_mask
else
mask
end
new_indices = mask |> Nx.flatten() |> Nx.cumulative_sum() |> Nx.subtract(1)
{new_vocab, removed_terms} =
Enum.reduce(vectorizer.vocabulary, {%{}, MapSet.new([])}, fn {term, old_index},
{vocab_acc, removed_acc} ->
case Nx.to_number(mask[old_index]) do
1 ->
{Map.put(vocab_acc, term, Nx.to_number(new_indices[old_index])), removed_acc}
_ ->
{vocab_acc, MapSet.put(removed_acc, term)}
end
end)
kept_indices = where_columns(mask)
if Nx.flat_size(kept_indices) == 0 do
raise "After pruning, no terms remain. Try a lower min_df or a higher max_df."
end
tf = Nx.take(tf, kept_indices, axis: 1)
{tf, new_vocab, removed_terms}
end
def _limit_features(self, X, vocabulary, high=None, low=None, limit=None):
if high is None and low is None and limit is None:
return X, set()
# Calculate a mask based on document frequencies
dfs = _document_frequency(X)
mask = np.ones(len(dfs), dtype=bool)
if high is not None:
mask &= dfs <= high if low is not none: mask &="dfs">= low
if limit is not None and mask.sum() > limit:
tfs = np.asarray(X.sum(axis=0)).ravel()
mask_inds = (-tfs[mask]).argsort()[:limit]
new_mask = np.zeros(len(dfs), dtype=bool)
new_mask[np.where(mask)[0][mask_inds]] = True
mask = new_mask
new_indices = np.cumsum(mask) - 1 # maps old indices to new
removed_terms = set()
for term, old_index in list(vocabulary.items()):
if mask[old_index]:
vocabulary[term] = new_indices[old_index]
else:
del vocabulary[term]
removed_terms.add(term)
kept_indices = np.where(mask)[0]
if len(kept_indices) == 0:
raise ValueError(
"After pruning, no terms remain. Try a lower min_df or a higher max_df."
)
return X[:, kept_indices], removed_terms
These functions show the most stark differences between the capabilities of NumPy versus those of Nx, as well as just the syntactic differences. The syntax of NumPy is much less verbose that that of Nx (especially considering that we are operating outside of a defn here which will inject its own implementation of the Kernel module to add custom operators), and there are just some capabilities in NumPy that are currently not possible in Nx. For example, in Nx you cannot do dynamic shape modifications like are done in the Python code new_mask[np.where(mask)[0][mask_inds]] = True, so I had to come up with other solutions that could achieve the same thing. Looking at the Python version, you realize that np.where(mask)[0] is only concerned with the resulting columns. This makes sense since each column represents a term in the vocabulary and each row represents a document in the corpus, so each item in a column represents the count of that term in that documents. So we are concerned with whole columns because term-frequency is calculated for each term, which again is represented by the whole column. So we can now use a combination of our function where_columns and Nx functions such as Nx.argsort, Nx.take, and Nx.multiply to rearrange the matrix such that items are sorted by our filter conditions, and then we can just take the number of items we want according to the supplied :limit.
It would take entirely too long for me to go over every difference between these two functions, but I implore you to look closely at these two implementations to gain a better understanding of how to convert NumPy code to Elixir Nx.
TFIDF Transformation
def fit(%__MODULE__{count_vectorizer: count_vectorizer} = vectorizer, corpus) do
{cv, tf} = CountVectorizer.fit_transform(count_vectorizer, corpus)
df = Scholar.Preprocessing.binarize(tf) |> Nx.sum(axes: [0])
idf =
if vectorizer.use_idf do
{n_samples, _n_features} = Nx.shape(tf)
df = Nx.add(df, if(vectorizer.smooth_idf, do: 1, else: 0))
n_samples = if vectorizer.smooth_idf, do: n_samples + 1, else: n_samples
Nx.divide(n_samples, df) |> Nx.log() |> Nx.add(1)
end
struct(vectorizer, count_vectorizer: cv, idf: idf)
end
def transform(%__MODULE__{count_vectorizer: count_vectorizer} = vectorizer, corpus) do
tf = CountVectorizer.transform(count_vectorizer, corpus)
tf =
if vectorizer.sublinear_tf do
Nx.select(Nx.equal(tf, 0), 0, Nx.log(tf) |> Nx.add(1))
else
tf
end
tf =
if vectorizer.use_idf do
unless vectorizer.idf do
raise "Vectorizer has not been fitted yet. Please call `fit_transform` or `fit` first."
end
Nx.multiply(tf, vectorizer.idf)
else
tf
end
tf =
case vectorizer.norm do
nil -> tf
norm -> Scholar.Preprocessing.normalize(tf, norm: norm)
end
tf
end
def fit_transform(%__MODULE__{} = vectorizer, corpus) do
vectorizer = fit(vectorizer, corpus)
{vectorizer, transform(vectorizer, corpus)}
end
class TfidfTransformer(
OneToOneFeatureMixin, TransformerMixin, BaseEstimator, auto_wrap_output_keys=None
):
def __init__(self, *, norm="l2", use_idf=True, smooth_idf=True, sublinear_tf=False):
self.norm = norm
self.use_idf = use_idf
self.smooth_idf = smooth_idf
self.sublinear_tf = sublinear_tf
def fit(self, X, y=None):
X = self._validate_data(
X, accept_sparse=("csr", "csc"), accept_large_sparse=not _IS_32BIT
)
if not sp.issparse(X):
X = sp.csr_matrix(X)
dtype = X.dtype if X.dtype in FLOAT_DTYPES else np.float64
if self.use_idf:
n_samples, n_features = X.shape
df = _document_frequency(X)
df = df.astype(dtype, copy=False)
# perform idf smoothing if required
df += int(self.smooth_idf)
n_samples += int(self.smooth_idf)
# log+1 instead of log makes sure terms with zero idf don't get
# suppressed entirely.
idf = np.log(n_samples / df) + 1
self._idf_diag = sp.diags(
idf,
offsets=0,
shape=(n_features, n_features),
format="csr",
dtype=dtype,
)
return self
def transform(self, X, copy=True):
X = self._validate_data(
X, accept_sparse="csr", dtype=FLOAT_DTYPES, copy=copy, reset=False
)
if not sp.issparse(X):
X = sp.csr_matrix(X, dtype=np.float64)
if self.sublinear_tf:
np.log(X.data, X.data)
X.data += 1
if self.use_idf:
# idf_ being a property, the automatic attributes detection
# does not work as usual and we need to specify the attribute
# name:
check_is_fitted(self, attributes=["idf_"], msg="idf vector is not fitted")
# *= doesn't work
X = X * self._idf_diag
if self.norm is not None:
X = normalize(X, norm=self.norm, copy=False)
return X
@property
def idf_(self):
# if _idf_diag is not set, this will raise an attribute error,
# which means hasattr(self, "idf_") is False
return np.ravel(self._idf_diag.sum(axis=0))
@idf_.setter
def idf_(self, value):
value = np.asarray(value, dtype=np.float64)
n_features = value.shape[0]
self._idf_diag = sp.spdiags(
value, diags=0, m=n_features, n=n_features, format="csr"
)
class TfidfVectorizer(CountVectorizer):
def fit(self, raw_documents, y=None):
self._check_params()
self._warn_for_unused_params()
self._tfidf = TfidfTransformer(
norm=self.norm,
use_idf=self.use_idf,
smooth_idf=self.smooth_idf,
sublinear_tf=self.sublinear_tf,
)
X = super().fit_transform(raw_documents)
self._tfidf.fit(X)
return self
def fit_transform(self, raw_documents, y=None):
self._check_params()
self._tfidf = TfidfTransformer(
norm=self.norm,
use_idf=self.use_idf,
smooth_idf=self.smooth_idf,
sublinear_tf=self.sublinear_tf,
)
X = super().fit_transform(raw_documents)
self._tfidf.fit(X)
# X is already a transformed view of raw_documents so
# we set copy to False
return self._tfidf.transform(X, copy=False)
def transform(self, raw_documents):
check_is_fitted(self, msg="The TF-IDF vectorizer is not fitted")
X = super().transform(raw_documents)
return self._tfidf.transform(X, copy=False)
I would consider this the most straight-forward translation example between the two implementations. One noticeable difference is that Nx.log doesn't handle zero values the same way NumPy does. While NumPy essentially ignores zeroes, Nx.log will throw a divide by zero error, so I use Nx.select to selectively ignore zero values, and only apply Nx.log to non-zero values. Additionally, I use Nx.multiply(tf, vectorizer.idf) to achieve the same thing as X = X * self._idf_diag as there is no need to construct a diagonal matrix since Nx.multiply broadcasts.
Conclusion
I would have liked to go into more detail for each example, but I think the code does a good job by itself showing the differences and steps required to translate a NumPy implementation to Nx. I think these examples illustrate how NumPy can obscure what operations are happening in an effort to make a more concise syntax, whereas some might consider Nx overly verbose in comparison. The more you familiarize yourself with both APIs, the better you will be able to identify places where you can do direct translation and places where you might have to be more creative.
Comment below with your own examples or if you have any other Python snippets you want to see converted to Elixir Nx!



